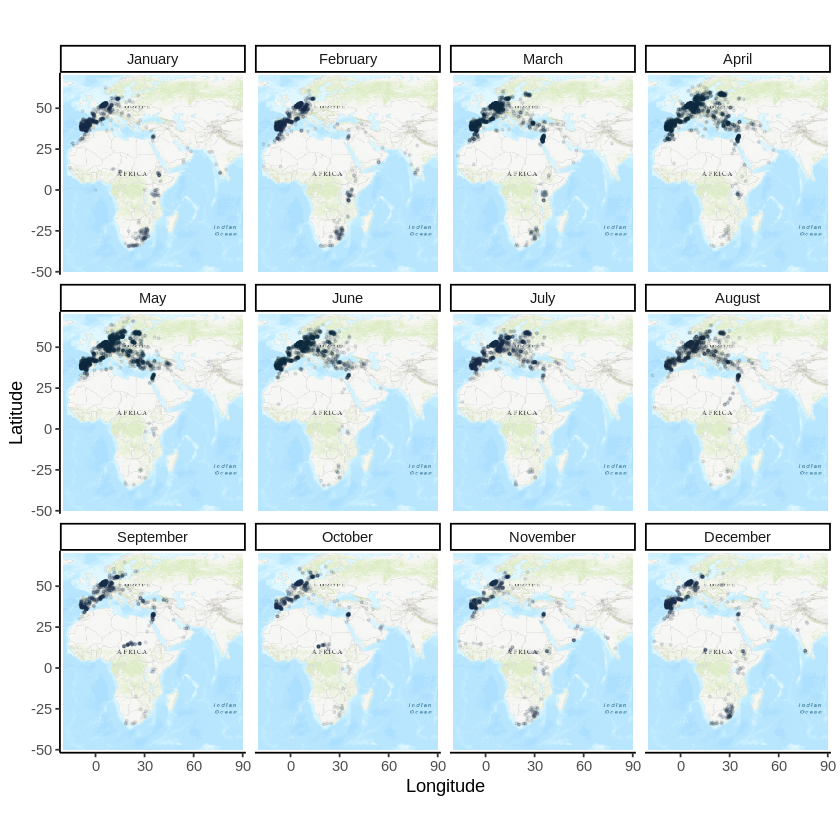
In a previous post, I discussed how to plot occurrence data from GBIF on a map. In this post, I will discuss how to plot a bird migration by producing an occurrence map for each month of the year. I will use the migration of the stork (Ciconia ciconia) as an example.
These are the libraries I will use for drawing the maps using OpenStreetMap data:
library(OpenStreetMap)
library(ggplot2)
library(sp)
library (rgdal)
library(readr)
Storks migrate from Europe where most of them breed, to southern Africa and India. The area of the map is therefore delimited by these coordinates:
LAT1 = 70 ; LAT2 = -50
LON1 = -20 ; LON2 = 90
Get the map from OpenstreetMap. A more detailed explanation of the parameters can be found here.
map <- openmap(
c(LAT2,LON1), c(LAT1,LON2),
zoom = 2,
type = "esri-topo",
mergeTiles = TRUE)
print("done loading map")
The map can now be drawn. Here, I used a longitude / latitude projection for simplicity. If you were to plot a distribution instead of occurrences, an equal area projection would be a better pick. Projection definitions can be found on the EPGS website.
# the definition of the projection proj <- "+proj=longlat +ellps=clrk66" # project the map map.proj <- openproj(map, projection = proj) # plot the map autoplot(map.proj) + theme_classic() + labs(x="longitude", y="latitude")
Get occurrence data from GBIF, as discussed here. Save the data, without unpacking it, to file. I stored it in a file called “data/GBIF_Ciconia_ciconia.zip”. The occurrence data can now be loaded into a variable. Note that it is not necessary to unzip the file before reading it, and that the data is in tab-separated format (TSV).
The data can now be loaded and cleaned. Optionally, as the data set has 600.000 entries, I chose to sub-sample it to speed things up.
# load the file into a variable
occurrences <- read_tsv("data/GBIF_Ciconia_ciconia.zip")
# Cleanup the data
occurrences.clean <- occurrences[
!is.na(occurrences$decimalLatitude) &
!is.na(occurrences$decimalLongitude) &
!is.na(occurrences$month),]
# randomly sample 25.000 entries
occurrences.sampled <-
occurrences.clean[sample(nrow(occurrences.clean), 25000),]
Now project the data using the same projection definition as for the map
coordinates(occurrences.sampled) <-
c("decimalLongitude", "decimalLatitude")
proj4string(occurrences.sampled) <- CRS(proj)
occurrences.proj <-
spTransform(occurrences.sampled, CRS=CRS(proj))
The map and the data can now be plotted, using facets to create a map for each month of the year
# return the name of the month
month_labeller <- function(variable,value){
return(month.name[value])
}
# plot the map and the data
autoplot(map.proj) +
geom_count(
data = data.frame(occurrences.proj),
aes(x = decimalLongitude, y = decimalLatitude,
color = ..prop..
), alpha=0.1, size=0.5, show.legend = FALSE
) +
scale_fill_gradient(low="red", high="yellow") +
facet_wrap(~month, labeller = month_labeller) +
theme_classic() +
labs(x="Longitude", y="Latitude")
References
GBIF.org (07 September 2020) GBIF Occurrence Download https://doi.org/10.15468/dl.vm74xx